The group is run by Hans-Petter Hersleth and Marta Hammerstad.
We are also a UiO Structural Biology Core Facilities node, so if you have questions or need help with structural biology you can contact us.
Research interest:
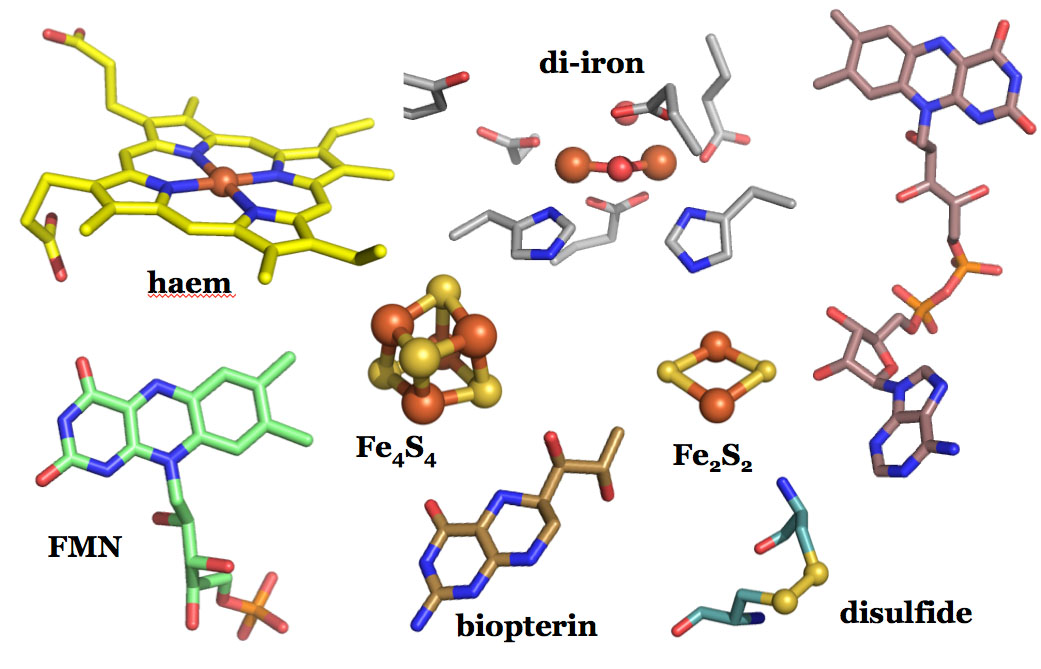
We have a general interest in the structure and function of proteins, especially redox enzymes using different cofactors to perform their functions. These include haem and other metalloproteins, flavin proteins and thiol-based proteins.
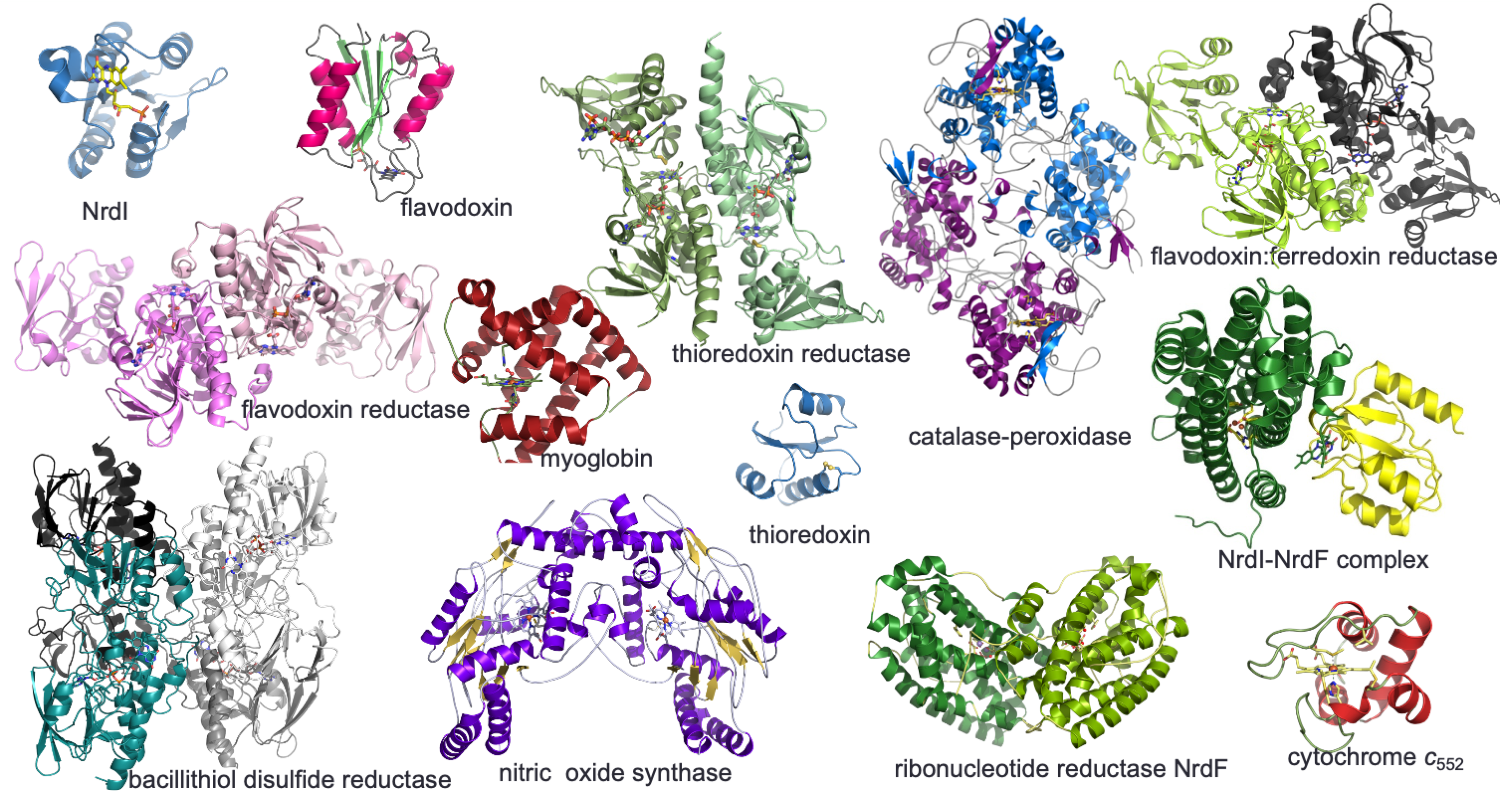
Redox enzymes are often part of redox networks, where other cofactor-based proteins activate these enzymes. A research focus is to decipher the enzyme mechanisms and activation networks in key redox enzyme systems:
-
Low molecular weight (LMW) thiols are important biological antioxidants. Many Firmicutes use the LMW thiol bacillithiol as a defense mechanism to buffer the intracellular redox environment and counteract oxidative stress encountered by human neutrophils during infections. One way of protection is through bacillithiolation of the redox active cysteins. This involves a network of several redox enzymes.
-
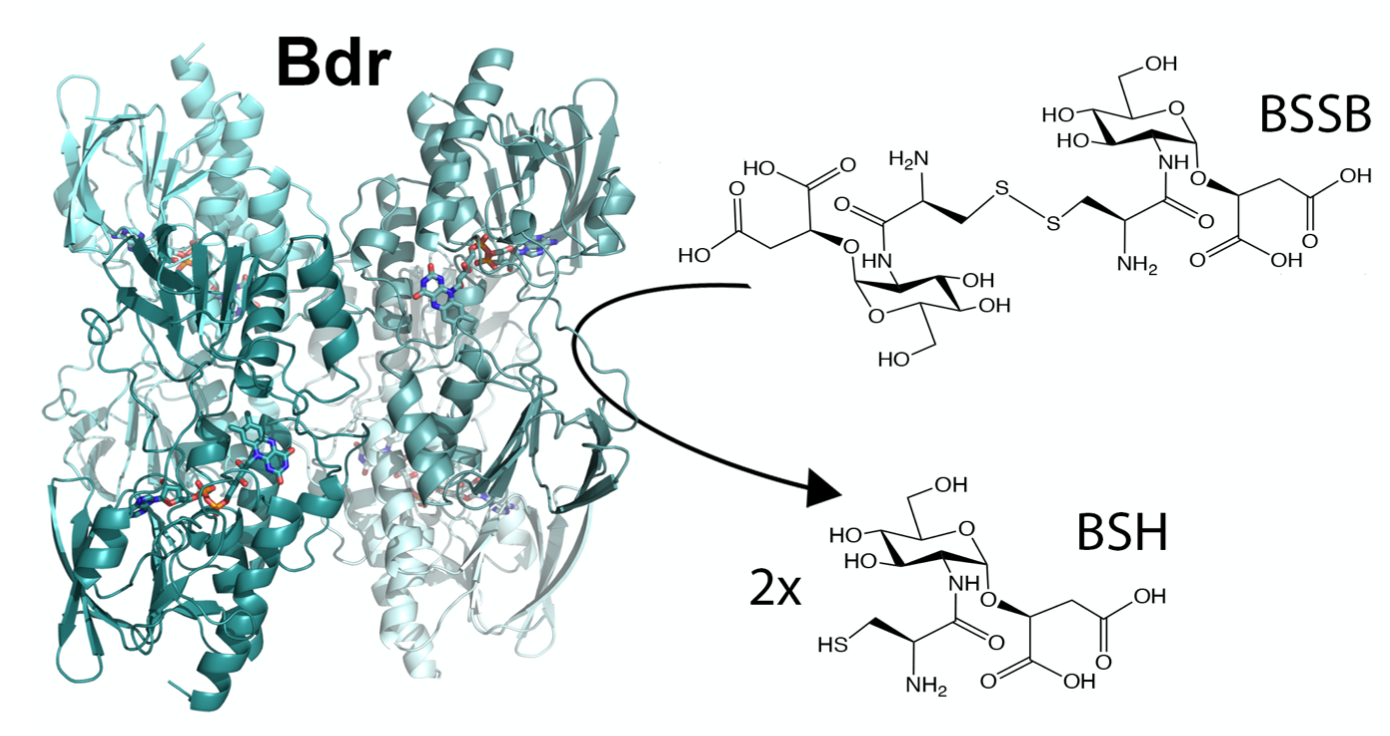
Structure of the protein that reduces oxidised bacillithiol after antioxidant defence by bacillithiol (Illustration: Hersleth, UiO) -
Ribonucleotide reductase (RNR) converts the RNA building blocks to DNA building blocks, and is one enzyme system we are focusing on. The activation of this system is different in humans and bacteria, as well as within the bacterial classes. These systems involve both di-iron, di-manganese, iron-sulfur, flavin and thiol-based proteins.
-

Reaction scheme of bacterial ribonucleotide reductase (Illustration: Hersleth, UiO) -
Another focus is on a more recently discovered group of thioredoxin reductase-like FAD-containing NADPH-dependent oxidoreductases in Firmicutes. Here, the individual enzymes has evolved to perform different redox reactions, where much of the detailed understanding is lacking.
-
A focus on haem proteins is on the high-valent reaction intermediates generated in several redox cycles of haem proteins.
- The X-rays used in protein crystallography can lead to radiation induced damage and reduction of the cofactor redox states in the protein crystals. To understand and cope with these effects, we are combining protein crystallography with in situ single crystal spectroscopic methods at synchrotrons. We have now a new project where we will combine this with neutron protein crystallography.
To decipher the functions of these redox proteins on a molecular and electronic level a combination of biochemical and biophysical methods are needed. We therefore combine protein crystallography (X-ray and neutron), different spectroscopic techniques both in solution and crystals (e.g. UV-vis, Raman), binding studies, kinetic studies, potentiometric studies and activity studies.
More info about the whole PX-Oslo core facilities.![]()
Funding

