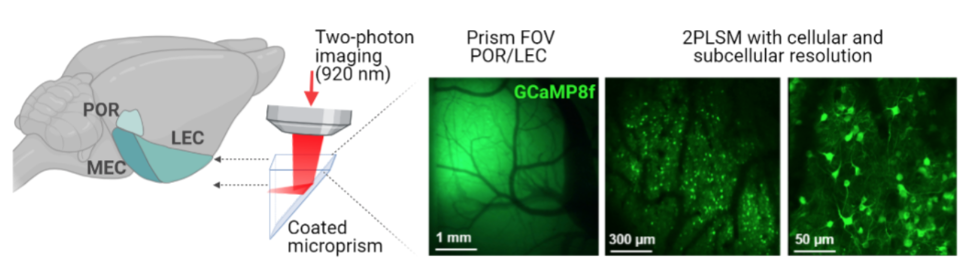
Mammals show remarkable flexibility of behaviors, seamlessly transferring knowledge from one domain for new learning in another domain. This ability called transfer learning relies on abstracting structures of the environment and generalizing prior experience to novel situations. Compared to the brain, transfer learning is a major challenge for AI systems that often need substantial re-training when presented to novel conditions. Thus, deep insight into the neural basis for transfer learning in the brain can inspire novel AI systems with potential excelled performance. Moreover, the brain regions essential to transfer learning are affected early in Alzheimer’s disease. Thus, understanding these neural circuitries holds the potential to explain the neural basis for early cognitive decline in this disease.
In our laboratory, we measure the activity of large neuron populations in animals performing learning tasks. By targeted interventions of neuron activity, we can identify critical elements of the neural network responsible for transfer learning in the brain.
The complexity of the resulting data requires new means to understand them. The candidate will work alongside experimentalists and develop tools to analyse and understand the neural dynamics. Moreover, the goal is to implement these insights into artificial neural networks and explore neural population dynamics in novel ways to eventually ignite brain-inspired AI systems. Based on the findings, new AI models will be developed and simulations will generate predictions that will be tested in the animal models for deep insight into brain function.
You will work in an interdisciplinary environment between physics, computer science and bioscience that combines experiments and modeling, providing a basis for new breakthroughs in our understanding of both artificial and biological neural networks.
The work includes collaboration with leading groups at Harvard University.
Requirements
- You must have a master degree in physics, computational neuroscience, artificial intelligence or similar.
- Documented experience from scientific computing or neural network simulations will be considered an advantage.
Supervisors
Professor Anders Malthe-Sørenssen
Call 2: Project start autumn 2022
This project is in call 2, starting autumn 2022.