Diabetes is a life-threatening chronic metabolic disease which impacts daily life and can have long term severe consequences for the patients due to serious secondary complications that can lead to premature death. Unfortunately, there is no cure for the disease yet, and therapy predominantly focuses on management to mimic the fine-tuning of glucose regulation in our body. Replacement therapies, based on introducing functional healthy insulin producing cells to patients requires donor material that is extremely sparse and requires tissue from deceased donors.
Induced pluripotent stem cells (iPS) can potentially differentiate into any cell type in the body. We have made progress in developing protocols for differentiating iPS cells into functional human insulin producing beta cells, but there is currently no protocol that can differentiate human iPS cells into fully glucose responsive cells.
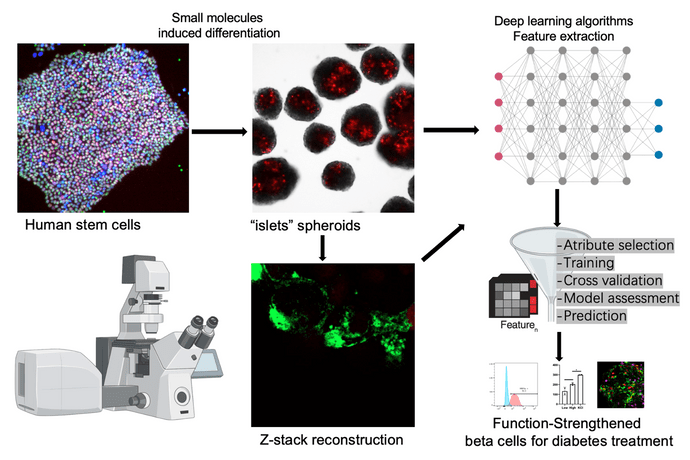
The dynamic process in which epigenetic, transcriptional, and metabolic changes lead to new cell identities is not fully understood. Likewise, the available tool to investigate these changes is based on time consuming and expensive assays measuring gene and protein expression that calls for innovative improvement. Image-based phenotypic screening relies on the extraction of multivariate information from cells cultured under a large variety of conditions. Technical advances in high-throughput microscopy enable screening in increasingly complex and biologically relevant model systems. To this end, this project will build on our recent observations that spatial relationships in the differentiating cell colonies are important for the final fate of the iPSC.
The project will use frequent, systematic live cell imaging with a range of fluorescent reporters during iPSC differentiation into true insulin producing cells. Deep-learning approaches will be used to analyze the spatial relationships in the images together with gene and protein expression data. Interpretation of traits of the trained network will be used to choose key changes to the protocol. The goal of the project is to optimize and improve differentiation efficiency and functionality to achieve self-organizing systems that work towards in vitro iPSC-derived beta-cell maturation.
Requirements
- The candidate must be motivated to develop both experimental and numerical tools for the project and have a MSc in physics, mathematics, computer science or a MSc in life science with experience in scientific programming.
- Candidates with documented experience in scientific programming, machine learning and live cell imaging will be prioritized.
Supervisors
Senior Researcher Hanne Scholtz
Call 2: Project start autumn 2022
This project is in call 2, starting autumn 2022.