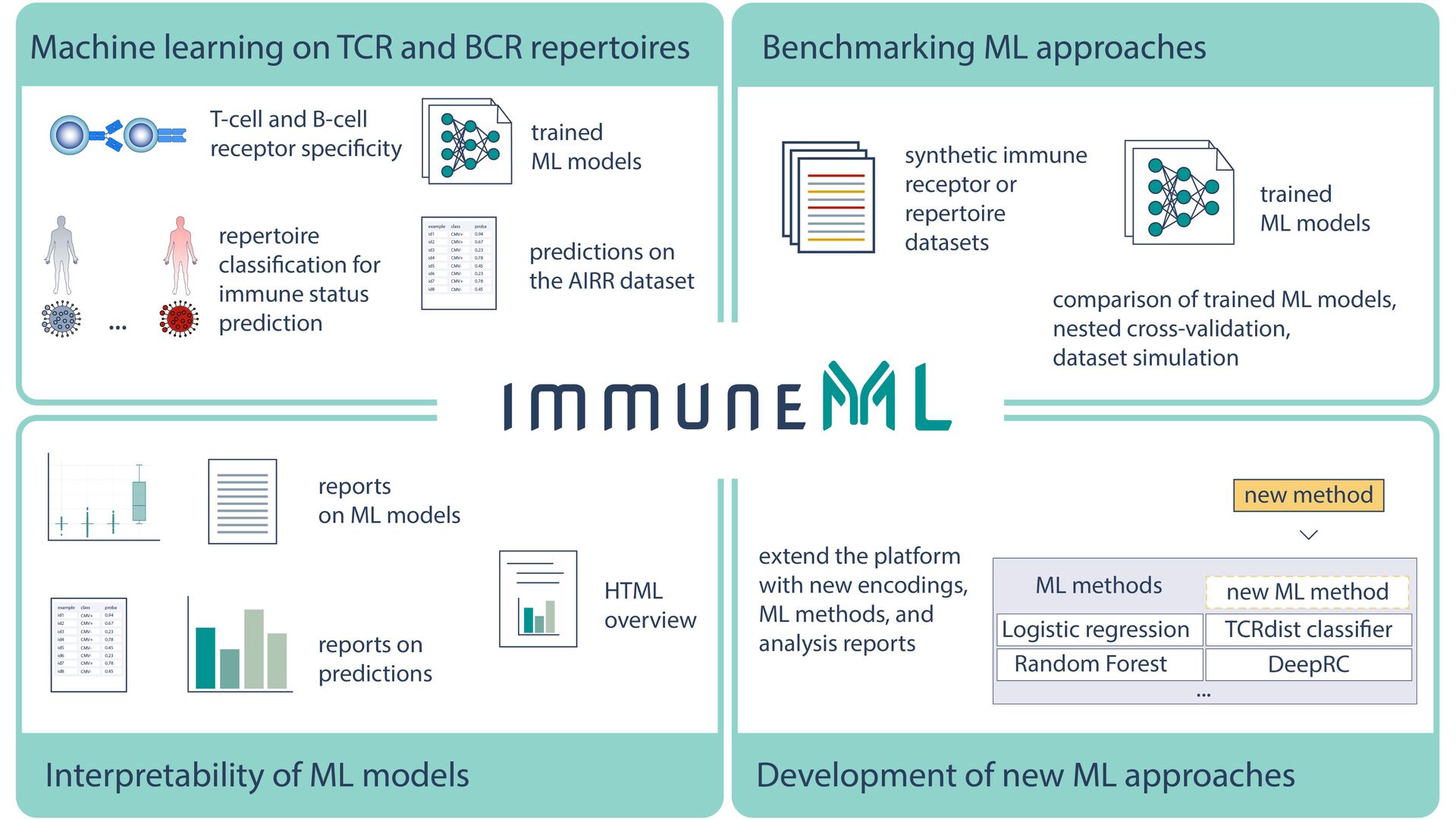
Adaptive immune receptor repertoires (AIRRs), including the receptors of B and T cells, reflect both past and current immune state. To this end, using machine learning (ML) on AIRRs is ideal for the application of AIRRs in diagnostic and therapeutic discoveries, as ML can identify complex sequence patterns. However, there is currently a paucity in the adoption of AIRR ML, due to challenges related to reproducibility, transparency, and interoperability.
To overcome these limitations, a multidisciplinary collaboration at the University of Oslo, including several PharmaTox-affiliated researchers, has realised an open-source software ecosystem: “immuneML”. The software is accessible to users, clinicians and researchers alike, to promote the adoption of AIRR-based diagnostic and therapeutic discoveries.
Read more about “immuneML” in Nature Machine Intelligence, and try out “immuneML” here. Remember to visit “immuneML” on Twitter!