Swarm v3: towards tera-scale amplicon clustering
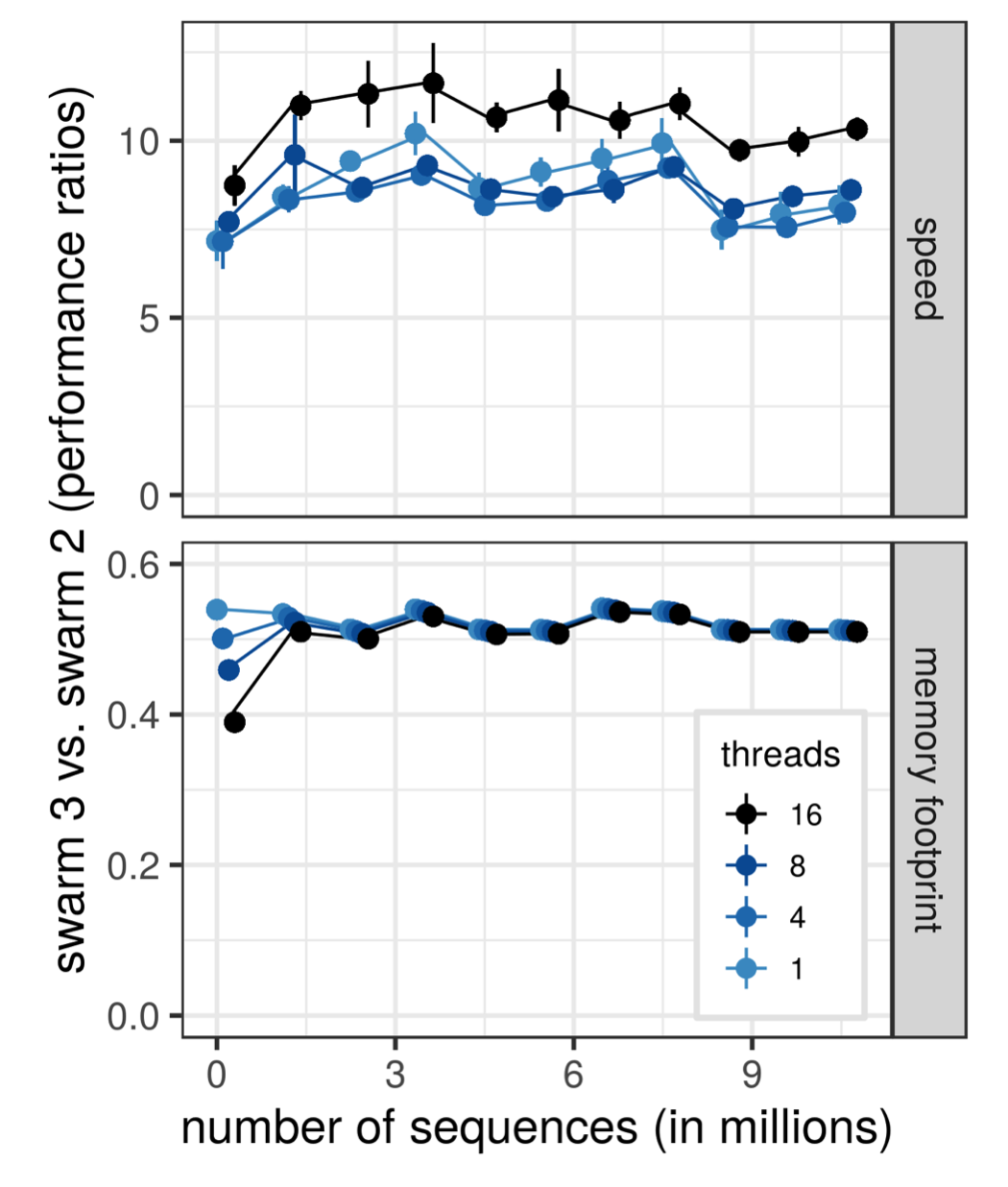
"The new version 3 of the Swarm software is a major step towards clustering extremely large amplicon datasets used in microbiome studies. It includes extensive improvements that make it about 10 times faster and twice as memory efficient as the previous version. Swarm makes it possible to cluster a huge dataset in reasonable time at one go, unlike other approaches where a large dataset needs to analysed in smaller portions," says Torbjørn Rognes, professor at the Centre for Bioinformatics.
The work was carried out in an international collaboration and was recently published in the journal Bioinformatics. The open-source tool is freely available on GitHub.
"These datasets are produced in studies to unravel ecological and evolutionary patterns within and across microbiomes using a kind of DNA barcode. Swarm is used to group similar barcode sequences together and classify them into different species. The algorithms developed for Swarm were based on ideas from several members of the team. We applied some very interesting computer science concepts, including a special hash function, to make the clustering so fast. The original version of Swarm was published in 2014 and since then we have also increased the code quality to make the software more robust. The code has been modernized and we have applied various automatic tests and quality checks to improve it. We hope it will be useful for the microbiome community."
Published Aug. 16, 2021 2:43 PM
- Last modified Aug. 16, 2021 3:14 PM
